Methods
Ecologists are focussing increasingly on explaining the structure of communities by enriching traditional food web data with additional information. Many community studies collect either the food web, species' body masses, or abundance data, and an ever increasing number of studies measure two or three of these data types and/or additional data. Various combinations of data allow different properties to be explored and different hypotheses to be tested.
Cheddar provides many relevant published methods. The examples below illustrate the different views of community data that contain different combinations of trophic links, body masses, and abundance data, following Cohen et al 2003, without going into detail. Most examples use the dataset of Tuesday Lake sampled in 1984 (Cohen et al 2003; Jonsson et al 2005).
| Trophic links | Body mass | Numerical abundance | Description |
|---|---|---|---|
| ✓ | Rank of body mass | ||
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotMvRankM(TL84)
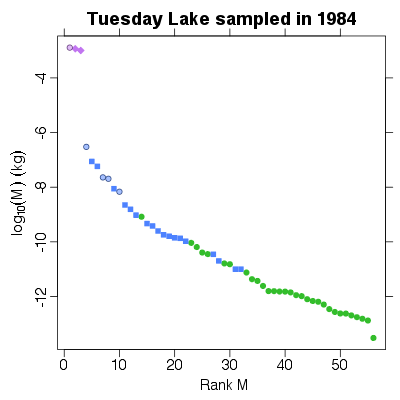
> PlotMvRankM(TL84, show.nodes.as="labels")
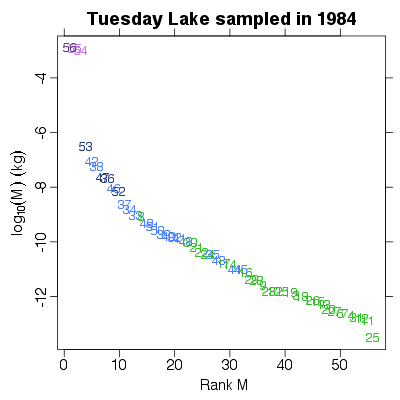
|
|||
| ✓ | Distribution of body mass | ||
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotMDistribution(TL84)
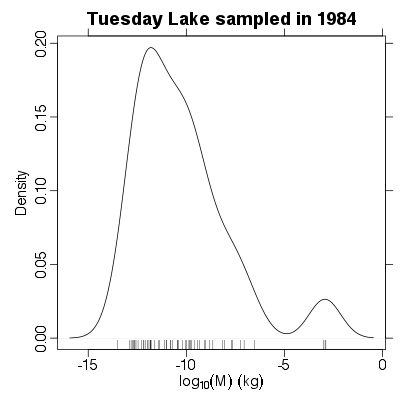
|
|||
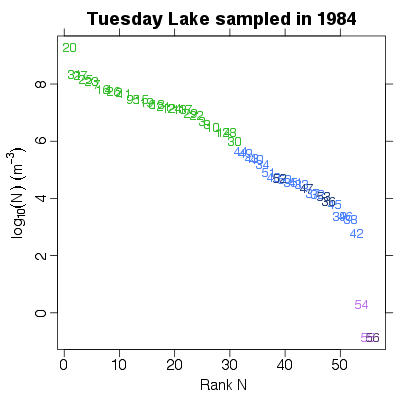
| ✓ | Rank of numerical abundance | ||
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotNvRankN(TL84)
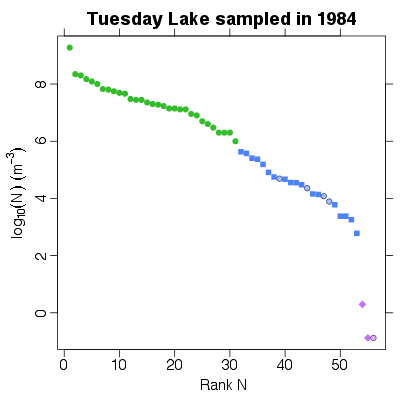
> PlotNvRankN(TL84, show.nodes.as="labels")
|
|||
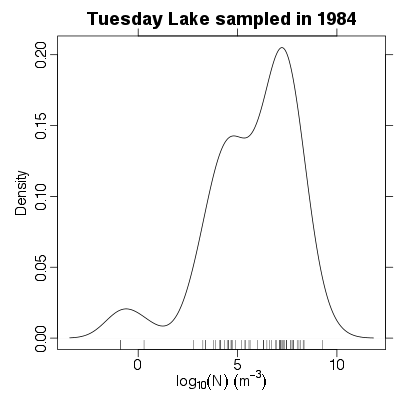
| ✓ | Distribution of numerical abundance | ||
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotNDistribution(TL84)
|
|||
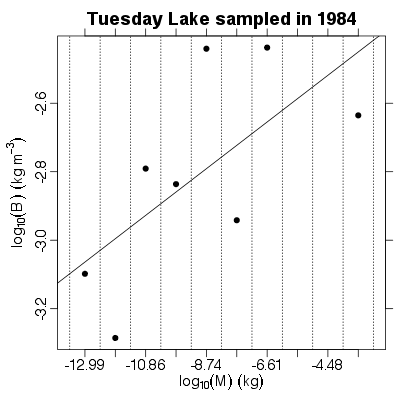
| ✓ | ✓ | Approximate biomass abundance spectrum | |
|
Functions:
References: Kerr & Dickie 2001; Examples > PlotBSpectrum(TL84)
|
|||
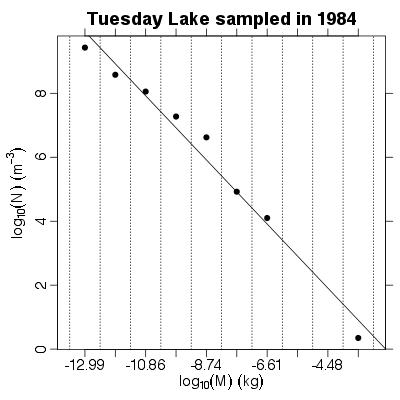
| ✓ | ✓ | Approximate numerical abundance spectrum | |
|
Functions:
References: Kerr & Dickie 2001; Examples > PlotNSpectrum(TL84)
|
|||
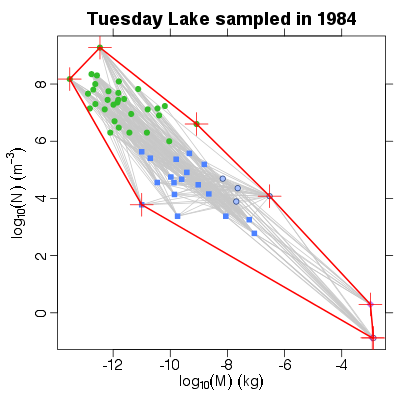
| ✓ | ✓ | Constraint space defined by a convex hull | |
|
Functions:
References: Leaper & Raffaelli 1999; Examples > ch <- NvMConvexHull(TL84)
> print(ch$area)
[1] 30.68266
> # Show the hull graphically
> PlotNvM(TL84)
> polygon(ch$points, lwd=2, border='red')
> points(ch$points, pch=3, col='red', cex=3)
|
|||
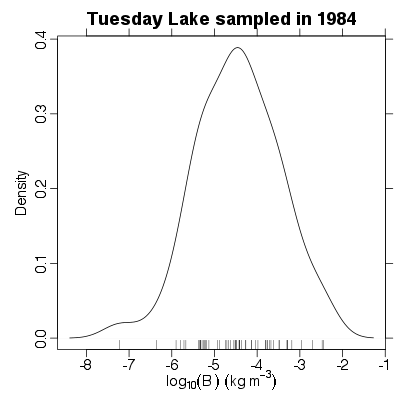
| ✓ | ✓ | Distribution of biomass abundance | |
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotBDistribution(TL84)
|
|||
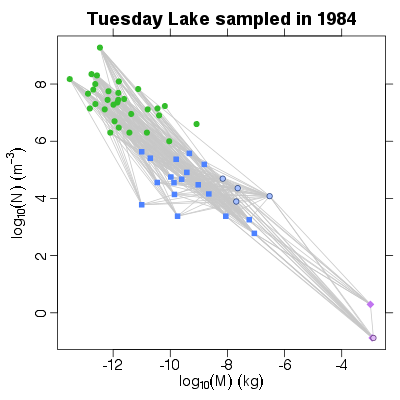
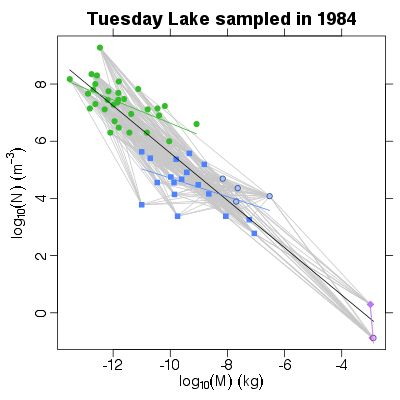
| ✓ | ✓ | Mass-abundance allometry | |
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotNvM(TL84)
> # Fit linear regressions to the whole community and to each group
> PlotNvM(TL84)
> models <- NvMLinearRegressions(TL84)
> PlotLinearModels(models)
<NA> producer invertebrate vert.ecto
"black" "#33bd2b" "#4d82ff" "#c175f0"
|
|||
| ✓ | ✓ | Rank of biomass abundance | |
|
Functions:
References: Cohen et al 2003; Jonsson et al 2005; Examples > PlotBvRankB(TL84)
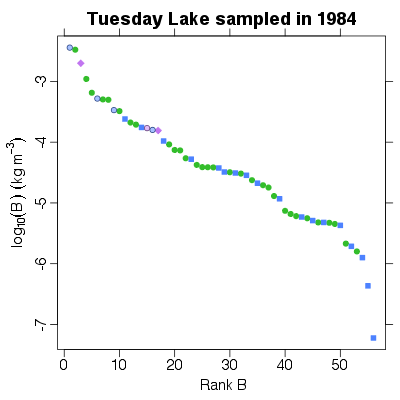
> PlotBvRankB(TL84, show.nodes.as="labels")
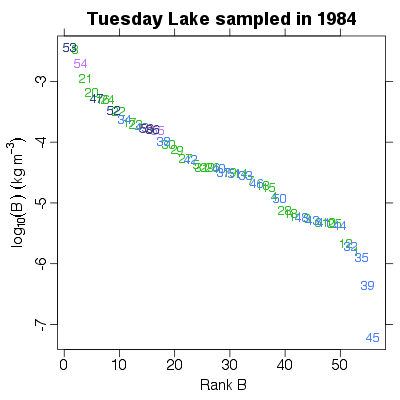
|
|||
| ✓ | Basic network properties | ||
|
Functions:
References: Martinez 1991; Cohen et al 2003; Jonsson et al 2005; Examples > print(NumberOfTrophicLinks(TL84))
[1] 269
> print(LinkageDensity(TL84))
[1] 4.803571
> print(DirectedConnectance(TL84))
[1] 0.08577806
|
|||
| ✓ | Feeding interactions as a matrix | ||
|
Functions:
Examples > PlotPredationMatrix(TL84)
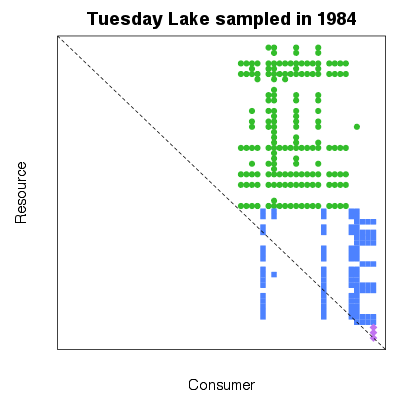
> pm <- PredationMatrix(TL84)
> # A square matrix
> print(class(pm))
[1] "matrix"
> print(NumberOfNodes(TL84))
[1] 56
> print(nrow(pm))
[1] 56
> print(ncol(pm))
[1] 56
> # All values are 0 or 1. 1 indicates a feeding link
> print(unique(as.vector(pm)))
[1] 0 1
> # 269 trophic links
> print(NumberOfTrophicLinks(TL84))
[1] 269
> print(sum(pm))
[1] 269
|
|||
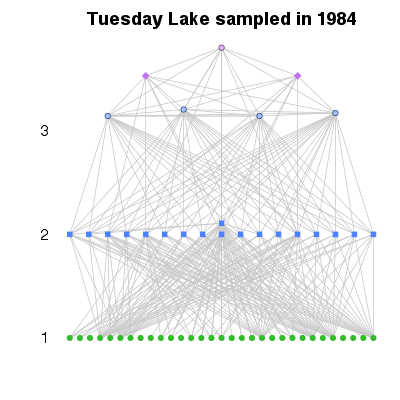
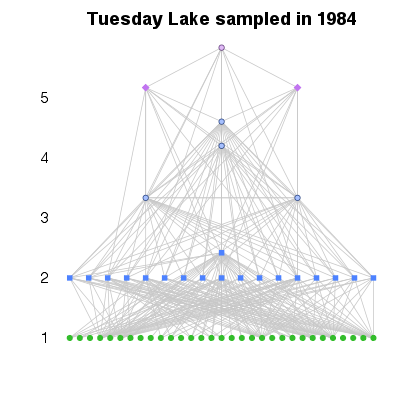
| ✓ | Food-web plotted vertically by trophic level | ||
|
Functions:
Examples > PlotWebByLevel(TL84)
> # A different measure of trophic level
> PlotWebByLevel(TL84, level='ChainAveragedTrophicLevel')
|
|||
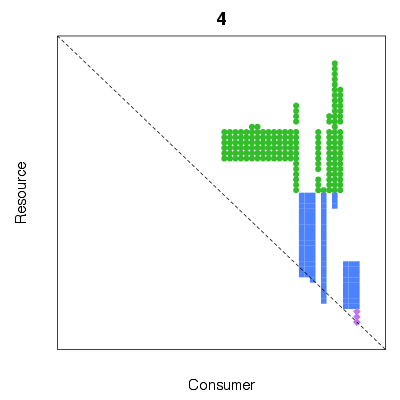
| ✓ | Intervality and diet gaps | ||
|
Functions:
References: Stouffer et al 2006; Zook et al 2011; Examples > # 5 repetitions of the simulated annealing method used to minimise diet gaps
> # Use the result with the smallest number of gaps
> res <- MinimiseSumDietGaps(TL84, n=5)
> print(res$sum.gaps)
[1] 4
> PlotPredationMatrix(res$reordered, main=res$sum.gaps)
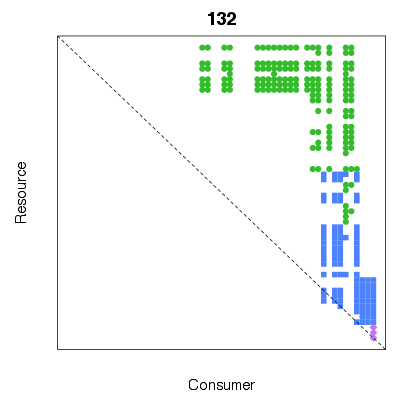
> # The data contain species' body masses - we can compare to number of diet
> # gaps when taxa are ordered by body mass
> byM <- OrderCommunity(TL84, 'M')
> print(SumDietGaps(byM))
[1] 132
> PlotPredationMatrix(byM, main=SumDietGaps(byM))
|
|||
| ✓ | Node connectivity categories | ||
|
Functions:
Examples > # A table of node connectivity
> connectivity <- NPS(TL84, c('IsBasalNode', 'IsIsolatedNode',
+ 'IsIntermediateNode', 'IsTopLevelNode'))
> print(connectivity)
IsBasalNode IsIsolatedNode IsIntermediateNode IsTopLevelNode
Nostoc sp. TRUE FALSE FALSE FALSE
Arthrodesmus sp. TRUE FALSE FALSE FALSE
Asterionella formosa FALSE TRUE FALSE FALSE
Cryptomonas sp. 1 TRUE FALSE FALSE FALSE
Cryptomonas sp. 2 TRUE FALSE FALSE FALSE
Chroococcus dispersus TRUE FALSE FALSE FALSE
Closteriopsis longissimus TRUE FALSE FALSE FALSE
Chrysosphaerella longispina FALSE TRUE FALSE FALSE
Dinobryon bavaricum TRUE FALSE FALSE FALSE
Dinobryon cylindricum TRUE FALSE FALSE FALSE
Dactylococcopsis fascicularis TRUE FALSE FALSE FALSE
Diceras sp. FALSE TRUE FALSE FALSE
Dictyosphaerium pulchellum TRUE FALSE FALSE FALSE
Dinobryon sertularia TRUE FALSE FALSE FALSE
Dinobryon sociale TRUE FALSE FALSE FALSE
Glenodinium quadridens TRUE FALSE FALSE FALSE
Microcystis aeruginosa TRUE FALSE FALSE FALSE
Mallomonas sp. 1 TRUE FALSE FALSE FALSE
Mallomonas sp. 2 TRUE FALSE FALSE FALSE
Unclassified flagellates TRUE FALSE FALSE FALSE
Peridinium limbatum TRUE FALSE FALSE FALSE
Peridinium cinctum TRUE FALSE FALSE FALSE
Peridinium pulsillum TRUE FALSE FALSE FALSE
Peridinium wisconsinense TRUE FALSE FALSE FALSE
Chromulina sp. TRUE FALSE FALSE FALSE
Rhizosolenia sp. FALSE TRUE FALSE FALSE
Selenastrum minutum TRUE FALSE FALSE FALSE
Spinocosmarium sp. FALSE TRUE FALSE FALSE
Staurastrum sp. FALSE TRUE FALSE FALSE
Synedra sp. TRUE FALSE FALSE FALSE
Trachelomonas sp. TRUE FALSE FALSE FALSE
Ascomorpha eucadis FALSE FALSE TRUE FALSE
Synchaeta sp. FALSE FALSE TRUE FALSE
Bosmina longirostris FALSE FALSE TRUE FALSE
Conochilus (solitary) FALSE FALSE TRUE FALSE
Cyclops varians rubellus FALSE FALSE TRUE FALSE
Diaphanosoma leuchtenbergianum FALSE FALSE TRUE FALSE
Daphnia pulex FALSE FALSE TRUE FALSE
Filinia longispina FALSE FALSE TRUE FALSE
Conochiloides dossuarius FALSE FALSE TRUE FALSE
Gastropus stylifer FALSE FALSE TRUE FALSE
Holopedium gibberum FALSE FALSE TRUE FALSE
Kellicottia sp. FALSE FALSE TRUE FALSE
Keratella cochlearis FALSE FALSE TRUE FALSE
Keratella testudo FALSE FALSE TRUE FALSE
Leptodiaptomus siciloides FALSE FALSE TRUE FALSE
Orthocyclops modestus FALSE FALSE TRUE FALSE
Ploesoma sp. FALSE FALSE TRUE FALSE
Polyarthra vulgaris FALSE FALSE TRUE FALSE
Trichocerca multicrinis FALSE FALSE TRUE FALSE
Trichocerca cylindrica FALSE FALSE TRUE FALSE
Tropocyclops prasinus FALSE FALSE TRUE FALSE
Chaoborus punctipennis FALSE FALSE TRUE FALSE
Phoxinus eos FALSE FALSE TRUE FALSE
Phoxinus neogaeus FALSE FALSE TRUE FALSE
Umbra limi FALSE FALSE FALSE TRUE
> # Each row sums to 1, confirming that a node fits only one of the four categories
> print(all(1==rowSums(connectivity)))
[1] TRUE
> # These summations are 1
> print(sum(FractionBasalNodes(TL84),
+ FractionIntermediateNodes(TL84),
+ FractionTopLevelNodes(TL84),
+ FractionIsolatedNodes(TL84)))
[1] 1
> print(sum(FractionConnectedNodes(TL84),
+ FractionIsolatedNodes(TL84)))
[1] 1
> print(sum(FractionBasalNodes(TL84),
+ FractionNonBasalNodes(TL84)))
[1] 1
> print(sum(FractionTopLevelNodes(TL84),
+ FractionNonTopLevelNodes(TL84)))
[1] 1
|
|||
| ✓ | Omnivory | ||
|
Functions:
References: Polis 1991; Examples > print(Omnivory(TL84))
[1] 0.1428571
|
|||
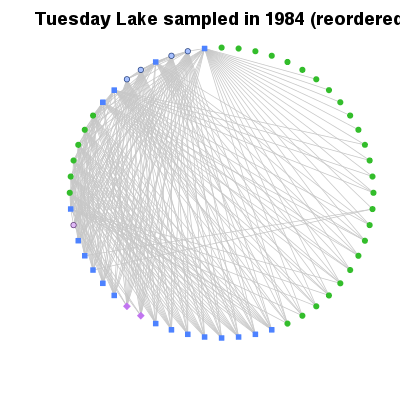
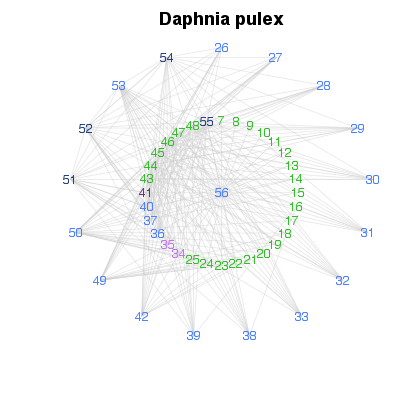
| ✓ | Overview of complexity | ||
|
Functions:
Examples > PlotCircularWeb(TL84)
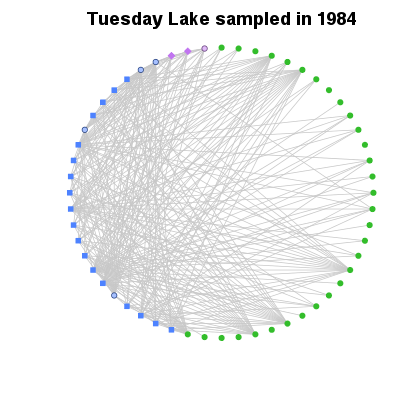
> # Order by increasing degree
> PlotCircularWeb(OrderCommunity(TL84, 'Degree'))
|
|||
| ✓ | Trophic level | ||
|
Functions:
References: Levine 1980; Cohen et al 2003; Jonsson et al 2005; Williams & Martinez 2004; Examples > PlotWebByLevel(TL84, level='PreyAveragedTrophicLevel')
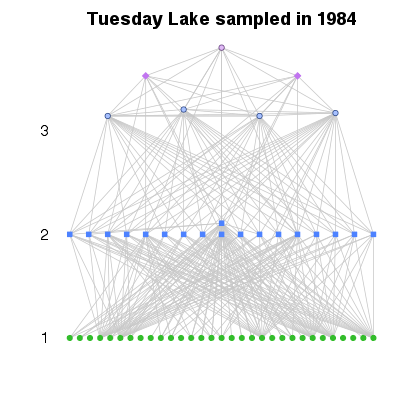
> PlotWebByLevel(TL84, level='ChainAveragedTrophicLevel')
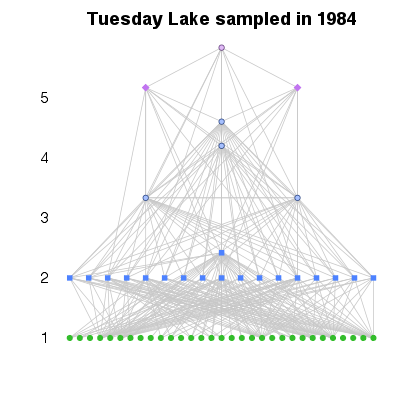
> # Six different measures of trophic level
> print(round(TrophicLevels(TL84), 2))
ShortestTL ShortWeightedTL LongestTL LongWeightedTL ChainAveragedTL PreyAveragedTL
Nostoc sp. 1 1.00 1 1.00 1.00 1.00
Arthrodesmus sp. 1 1.00 1 1.00 1.00 1.00
Asterionella formosa 1 1.00 1 1.00 1.00 1.00
Cryptomonas sp. 1 1 1.00 1 1.00 1.00 1.00
Cryptomonas sp. 2 1 1.00 1 1.00 1.00 1.00
Chroococcus dispersus 1 1.00 1 1.00 1.00 1.00
Closteriopsis longissimus 1 1.00 1 1.00 1.00 1.00
Chrysosphaerella longispina 1 1.00 1 1.00 1.00 1.00
Dinobryon bavaricum 1 1.00 1 1.00 1.00 1.00
Dinobryon cylindricum 1 1.00 1 1.00 1.00 1.00
Dactylococcopsis fascicularis 1 1.00 1 1.00 1.00 1.00
Diceras sp. 1 1.00 1 1.00 1.00 1.00
Dictyosphaerium pulchellum 1 1.00 1 1.00 1.00 1.00
Dinobryon sertularia 1 1.00 1 1.00 1.00 1.00
Dinobryon sociale 1 1.00 1 1.00 1.00 1.00
Glenodinium quadridens 1 1.00 1 1.00 1.00 1.00
Microcystis aeruginosa 1 1.00 1 1.00 1.00 1.00
Mallomonas sp. 1 1 1.00 1 1.00 1.00 1.00
Mallomonas sp. 2 1 1.00 1 1.00 1.00 1.00
Unclassified flagellates 1 1.00 1 1.00 1.00 1.00
Peridinium limbatum 1 1.00 1 1.00 1.00 1.00
Peridinium cinctum 1 1.00 1 1.00 1.00 1.00
Peridinium pulsillum 1 1.00 1 1.00 1.00 1.00
Peridinium wisconsinense 1 1.00 1 1.00 1.00 1.00
Chromulina sp. 1 1.00 1 1.00 1.00 1.00
Rhizosolenia sp. 1 1.00 1 1.00 1.00 1.00
Selenastrum minutum 1 1.00 1 1.00 1.00 1.00
Spinocosmarium sp. 1 1.00 1 1.00 1.00 1.00
Staurastrum sp. 1 1.00 1 1.00 1.00 1.00
Synedra sp. 1 1.00 1 1.00 1.00 1.00
Trachelomonas sp. 1 1.00 1 1.00 1.00 1.00
Ascomorpha eucadis 2 2.00 2 2.00 2.00 2.00
Synchaeta sp. 2 2.00 2 2.00 2.00 2.00
Bosmina longirostris 2 2.00 2 2.00 2.00 2.00
Conochilus (solitary) 2 2.00 2 2.00 2.00 2.00
Cyclops varians rubellus 3 3.07 4 3.57 3.33 3.14
Diaphanosoma leuchtenbergianum 2 2.00 2 2.00 2.00 2.00
Daphnia pulex 2 2.05 3 2.55 2.42 2.11
Filinia longispina 2 2.00 2 2.00 2.00 2.00
Conochiloides dossuarius 2 2.00 2 2.00 2.00 2.00
Gastropus stylifer 2 2.00 2 2.00 2.00 2.00
Holopedium gibberum 2 2.00 2 2.00 2.00 2.00
Kellicottia sp. 2 2.00 2 2.00 2.00 2.00
Keratella cochlearis 2 2.00 2 2.00 2.00 2.00
Keratella testudo 2 2.00 2 2.00 2.00 2.00
Leptodiaptomus siciloides 2 2.00 2 2.00 2.00 2.00
Orthocyclops modestus 3 3.10 5 4.10 4.20 3.21
Ploesoma sp. 2 2.00 2 2.00 2.00 2.00
Polyarthra vulgaris 2 2.00 2 2.00 2.00 2.00
Trichocerca multicrinis 2 2.00 2 2.00 2.00 2.00
Trichocerca cylindrica 2 2.00 2 2.00 2.00 2.00
Tropocyclops prasinus 3 3.07 4 3.57 3.33 3.14
Chaoborus punctipennis 2 2.59 6 4.59 4.60 3.17
Phoxinus eos 3 3.26 7 5.26 5.17 3.53
Phoxinus neogaeus 3 3.26 7 5.26 5.17 3.53
Umbra limi 3 3.40 8 5.90 5.84 3.80
|
|||
| ✓ | Trophic similarity | ||
|
Functions:
References: Williams & Martinez 2000; Examples > print(MeanMaximumTrophicSimilarity(TL84))
[1] 0.8520697
|
|||
| ✓ | Trophic generality and vulnerability | ||
|
Functions:
References: Williams & Martinez 2000; Examples > print(TrophicGenerality(TL84))
Nostoc sp. Arthrodesmus sp. Asterionella formosa Cryptomonas sp. 1 Cryptomonas sp. 2 Chroococcus dispersus
0 0 0 0 0 0
Closteriopsis longissimus Chrysosphaerella longispina Dinobryon bavaricum Dinobryon cylindricum Dactylococcopsis fascicularis Diceras sp.
0 0 0 0 0 0
Dictyosphaerium pulchellum Dinobryon sertularia Dinobryon sociale Glenodinium quadridens Microcystis aeruginosa Mallomonas sp. 1
0 0 0 0 0 0
Mallomonas sp. 2 Unclassified flagellates Peridinium limbatum Peridinium cinctum Peridinium pulsillum Peridinium wisconsinense
0 0 0 0 0 0
Chromulina sp. Rhizosolenia sp. Selenastrum minutum Spinocosmarium sp. Staurastrum sp. Synedra sp.
0 0 0 0 0 0
Trachelomonas sp. Ascomorpha eucadis Synchaeta sp. Bosmina longirostris Conochilus (solitary) Cyclops varians rubellus
0 6 6 11 7 16
Diaphanosoma leuchtenbergianum Daphnia pulex Filinia longispina Conochiloides dossuarius Gastropus stylifer Holopedium gibberum
14 28 6 7 6 19
Kellicottia sp. Keratella cochlearis Keratella testudo Leptodiaptomus siciloides Orthocyclops modestus Ploesoma sp.
6 6 6 16 17 6
Polyarthra vulgaris Trichocerca multicrinis Trichocerca cylindrica Tropocyclops prasinus Chaoborus punctipennis Phoxinus eos
6 6 6 16 22 9
Phoxinus neogaeus Umbra limi
9 12
|
|||
| ✓ | Trophic species | ||
|
Functions:
References: Briand & Cohen 1984; Pimm et al 1991; Jonsson et al 2005; Examples > TL84.lumped <- LumpTrophicSpecies(TL84)
> print(length(unique(TrophicSpecies(TL84)))) # 22 trophic species in TL84...
[1] 22
> print(NumberOfNodes(TL84.lumped)) # ... and 22 nodes in the lumped web
[1] 22
|
|||
| ✓ | Shortest trophic paths | ||
|
Functions:
Examples > PlotWagonWheel(TL84, 'Ploesoma sp.', main= 'Ploesoma sp.')
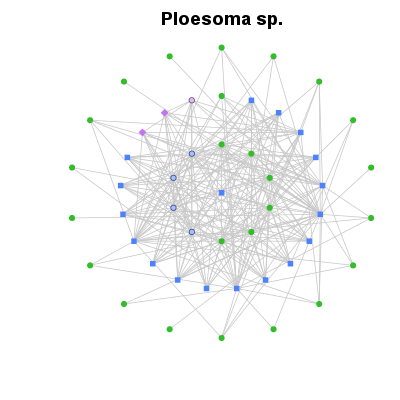
> # 'Daphnia pulex' is the focus, nodes ordered by degree (total number of
> # trophic links), lines partially transparent. The six isolated species are not shown.
> PlotWagonWheel(OrderCommunity(TL84, 'Degree'), 'Daphnia pulex',
+ show.nodes.as='labels', link.col=rgb(0.8,0.8,0.8,0.5),
+ main='Daphnia pulex')
|
|||
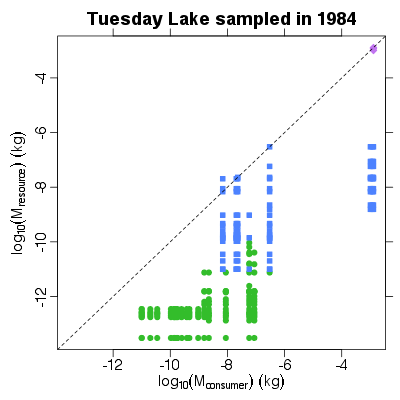
| ✓ | ✓ | Resource body mass against consumer body mass | |
|
Functions:
References: Jonsson et al 2005; Examples > PlotMRvMC(TL84)
|
|||
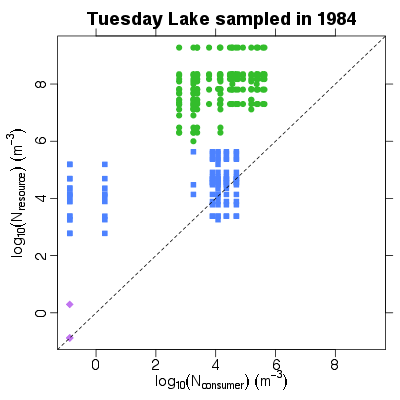
| ✓ | ✓ | Resource numerical abundance against consumer numerical abundance | |
|
Functions:
References: Jonsson et al 2005; Examples > PlotNRvNC(TL84)
|
|||
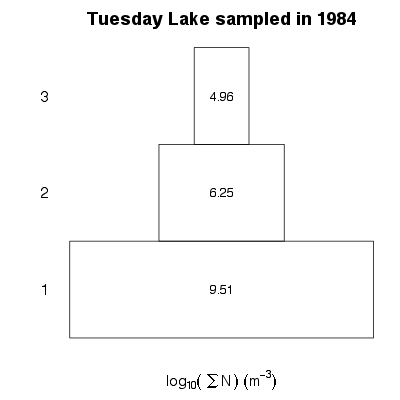
| ✓ | ✓ | Numerical abundance pyramid | |
|
Functions:
Examples > PlotNPyramid(TL84)
|
|||
| ✓ | ✓ | ✓ | Resource biomass abundance against consumer biomass abundance |
|
Functions:
Examples > PlotBRvBC(TL84)
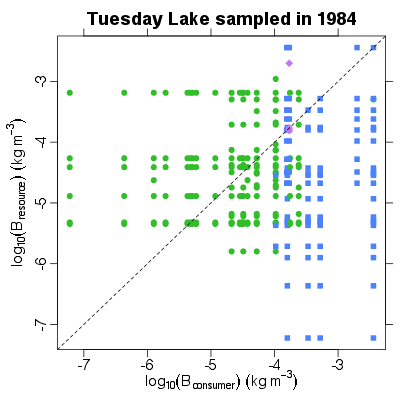
|
|||
| ✓ | ✓ | ✓ | Quantitative food-web descriptors based on information-theory indices |
|
Functions:
References: Bersier et al 2002; Examples > # Table 2 of Bersier et al 2002
> table <- QuantitativeDescriptors(ChesapeakeBay, 'biomass.flow')
> print(round(table, 2))
Qualitative Unweighted Weighted
Fraction top level 0.36 0.36 0.45
Fraction intermediate 0.48 0.48 0.39
Fraction basal 0.15 0.15 0.15
Ratio resources:consumers 0.75 0.71 0.32
Link density 2.21 1.37 2.10
Connectance 0.07 0.04 0.06
Fraction links top:intermediate 0.40 0.45 0.01
Fraction links top:basal 0.05 0.05 0.20
Fraction links intermediate:intermediate 0.29 0.29 0.11
Fraction links intermediate:basal 0.26 0.21 0.68
Mean chain length 3.16 1.60 1.52
Median chain length 3.00 1.62 0.67
SD chain length 1.10 0.49 1.91
Max chain length 6.00 2.97 6.41
Degree of omnivory 0.45 0.51 0.28
Generality 2.61 1.80 1.37
Vulnerability 3.48 1.91 2.82
SD standardised generality 0.74 0.65 1.88
SD standardised vulnerability 1.19 1.03 4.07
|
|||
| ✓ | ✓ | ✓ | Biomass abundance pyramid |
|
Functions:
Examples > PlotBPyramid(TL84)
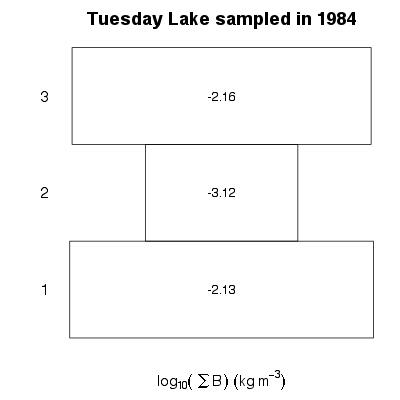
|
|||
| ✓ | ✓ | ✓ | Tri-trophic statistics |
|
Functions:
References: Cohen et al 2009; Examples > # Figure 1 D from Cohen et al 2009
> PlotAuppervAlower(TL84)
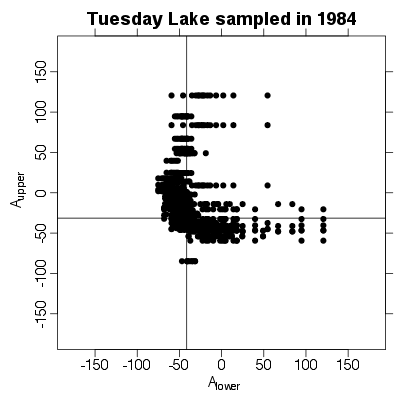
> # Table 1 from Cohen et al 2009
> collection <- CommunityCollection(list(TL84, TL86, YthanEstuary))
> table <- NvMTriTrophicTable(collection)
> print(round(table, 2))
Tuesday Lake sampled in 1984 Tuesday Lake sampled in 1986 Ythan Estuary
Mean link length 6.33 5.90 7.29
Mean L upper 5.41 3.43 5.06
Mean L lower 5.99 5.69 6.15
2 x mean link length 12.67 11.79 14.57
Mean 2-span 11.02 8.65 10.51
Mean L upper + L lower 11.40 9.12 11.20
2 x mean link length / mean 2-span 1.15 1.36 1.39
Mean L upper + L lower/ mean 2-span 1.03 1.05 1.07
Mean count chain length 4.84 4.84 4.43
Mean count chain length x mean link length 30.62 28.56 32.31
Community span 20.78 22.66 21.98
Mean count chain length x mean link length / community span 1.47 1.26 1.47
Mean sum chain lengths 19.96 23.33 16.88
Mean chain span 18.71 20.63 13.18
Mean chain span / community span 0.90 0.91 0.60
Mean sum chain lengths / mean chain span 1.07 1.13 1.28
Mean sum chain lengths / community span 0.96 1.03 0.77
L 264.00 236.00 379.00
S^2 2500.00 2601.00 8281.00
L/S^2 0.11 0.09 0.05
L/S 5.28 4.63 4.16
L 269.00 241.00 417.00
S^2 3136.00 3249.00 8464.00
L/S^2 0.09 0.07 0.05
L/S 4.80 4.23 4.53
|
|||
References
Bersier, L. and Banašek-Richter, C. and Cattin, M. (2002) Ecology 80 2394-2407
Williams, R.J. and Martinez, N.D. (2000) Simple rules yield complex food webs. Nature 404, 180-183
